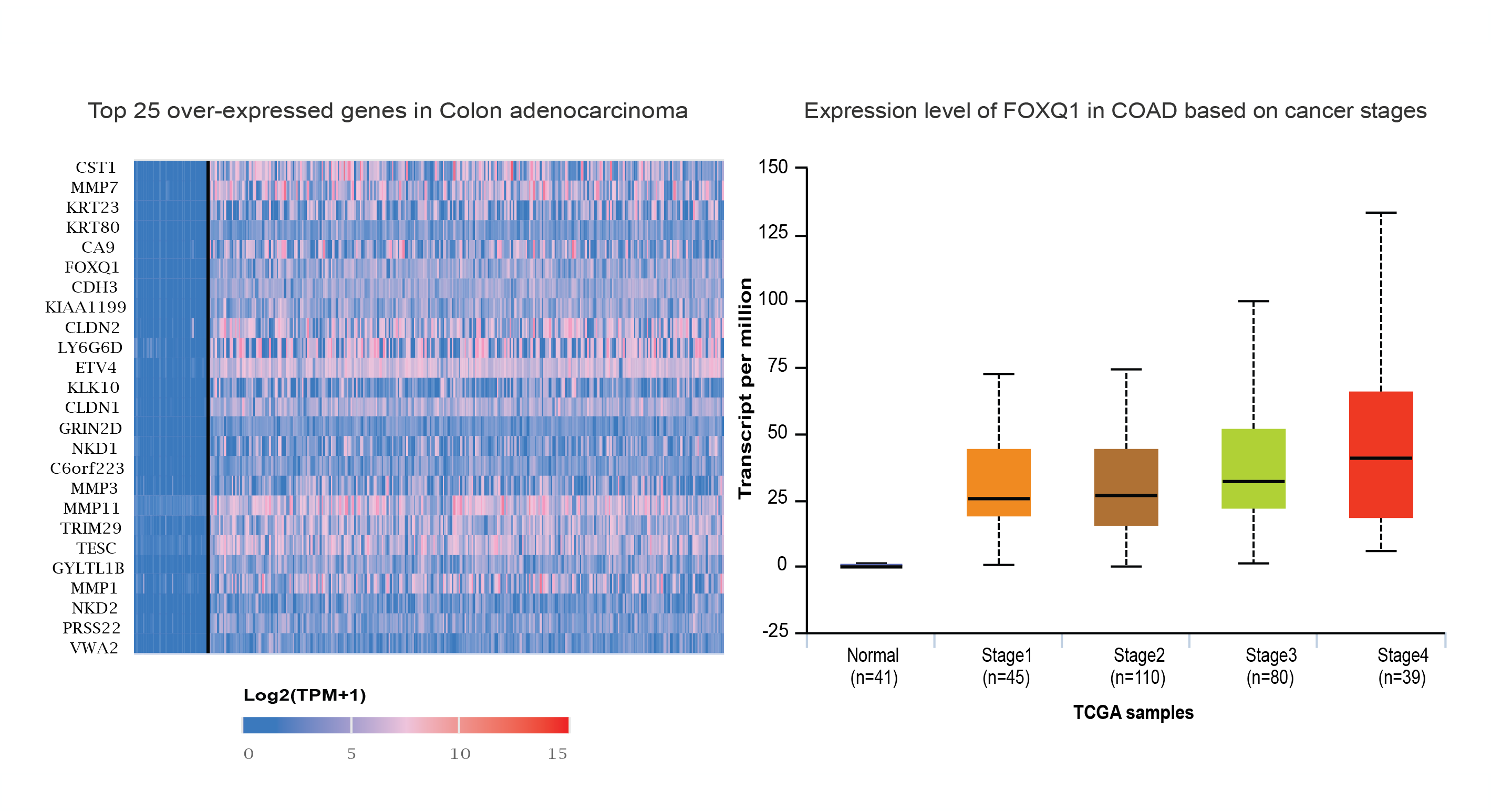
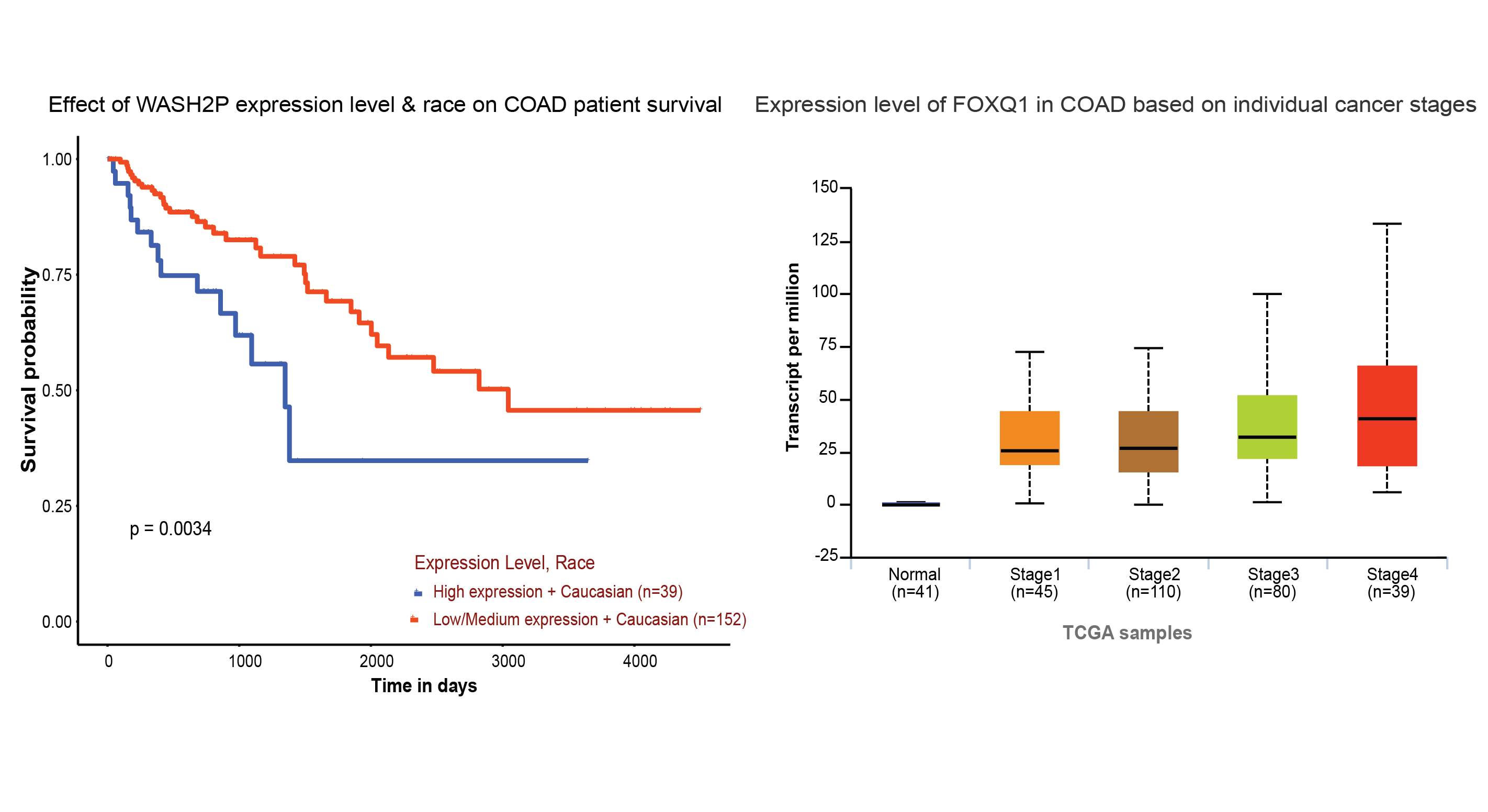
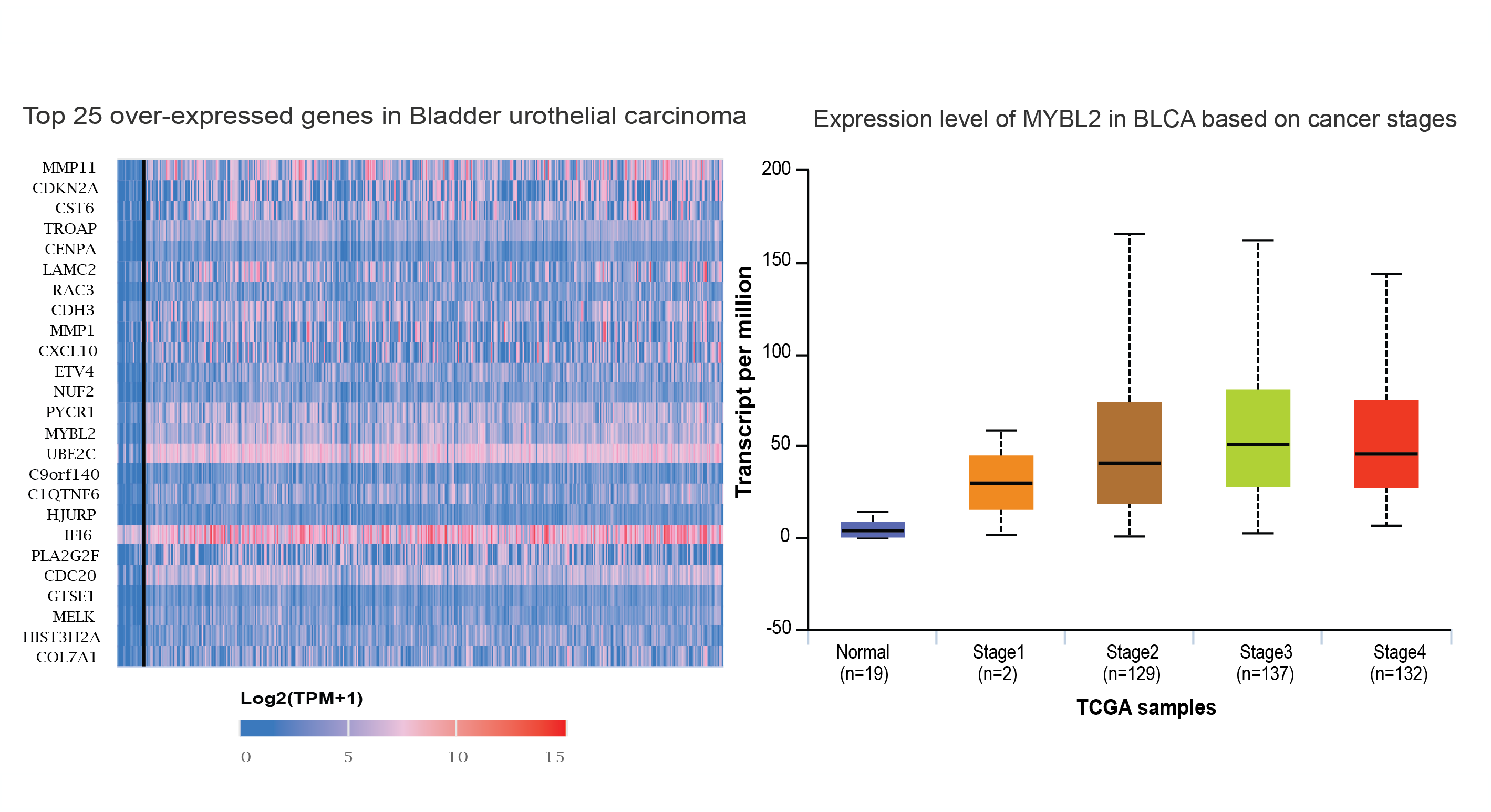
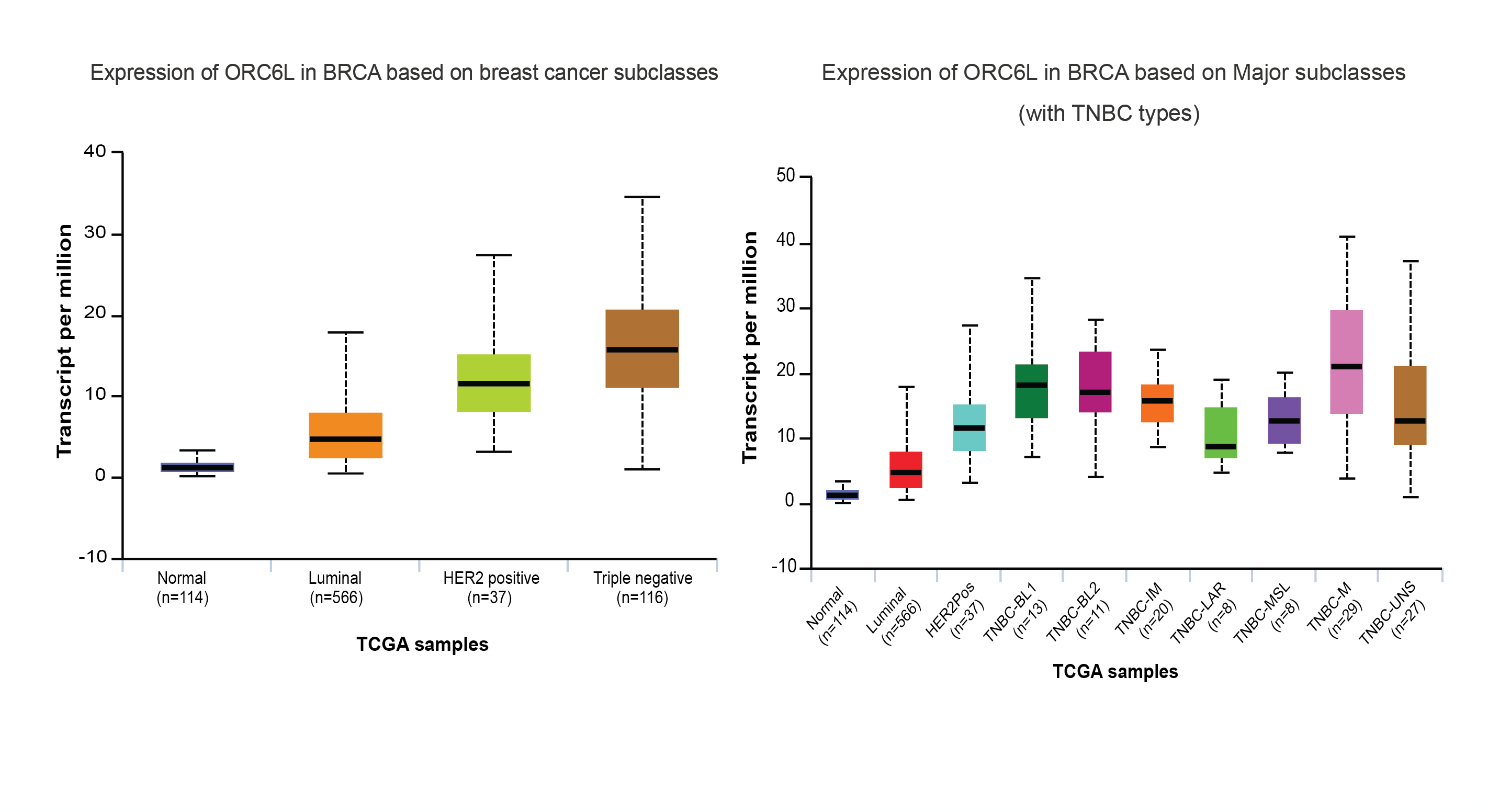
The University of ALabama at Birmingham CANcer data analysis Portal
UALCAN is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data. It is built on PERL-CGI with high quality graphics using javascript and CSS. UALCAN is designed to, a) provide easy access to publicly available cancer OMICS data (TCGA, MET500, CPTAC and CBTTC), b) allow users to identify biomarkers or to perform in silico validation of potential genes of interest, c) provide graphs and plots depicting expression profile and patient survival information for protein-coding, miRNA-coding and lincRNA-coding genes, d) evaluate epigenetic regulation of gene expression by promoter methylation, e) perform pan-cancer gene expression analysis, f) Provide additional information about the selected genes/targets by linking to GeneCards, Pubmed, TargetScan, The human protein atlas, DRUGBANK, Open Targets and the GTEx. These resources allow researchers to gather valuable information and data about the genes/targets of interest, g) provide clinical proteomic consortium data analysis including total/phospho-proteins and h) provide pediatric brain tumor gene expression and protein expression analysis.
Follow us on Twitter

Please cite:
1) Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022 Mar;25:18-27. doi: 10.1016/j.neo.2022.01.001 [PMID: 35078134]
2) Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Rodriguez IP, Chakravarthi BVSK and Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017 Aug;19(8):649-658. doi: 10.1016/j.neo.2017.05.002 [PMID:28732212]
Supported in part by NIH/NCI - Comprehensive Partnerships to Advance Cancer Health - 5U54CA118948
04/21/2025
Karthikeyan SK, Chandrashekar DS, Sahai S, Shrestha S, Aneja R, Singh R, Kleer CG, Kumar S, Qin ZS, Nakshatri H, Manne U, Creighton CJ, Varambally S. MammOnc-DB, an integrative breast cancer data analysis platform for target discovery. NPJ Breast Cancer. 2025 Apr 18;11(1):35. doi: 10.1038/s41523-025-00750-x.[PMID: 40251157]
02/12/2026
11/10/2023
09/14/2023
08/16/2022
05/13/2022
12/13/2021